Broken Timeline - Prologue Mac OS
I just upgraded to Mac OS X 10.5 Server (from Server 10.4.11) on a tiny 1.5Ghz 1GB Mac Mini. Before the OS upgrade, it ran a copy of Apache httpd v2.0.x, compiled from source, for pretty much uninterrupted for almost 3 years. I am now running the included version of httpd 2.2 (unmodified by me) after running the little 1.3 - 2.2 migration script. If you have a newer Mac, there is no physical option to install Mac OS versions older than your current Mac model. For instance, if your MacBook was released in 2014, don’t expect it to run any OS released prior of that time, because older Apple OS versions simply do not include hardware drivers for your Mac. Explore the world of Mac. Check out MacBook Pro, MacBook Air, iMac, Mac mini, and more. Visit the Apple site to learn, buy, and get support. Operating System macOS 10.13 High Sierra Apple MacBook Air MJVM2LL/A 11.6 Inch Laptop (Intel Core i5 Dual-Core 1.6GHz up to 2.7GHz, 4GB RAM, 128GB SSD, Wi-Fi, Bluetooth 4.0, Integrated Intel HD Graphics 6000, Mac OS) (Renewed).
- Broken Timeline - Prologue Mac Os Download
- Broken Timeline - Prologue Mac Os X
- Broken Timeline - Prologue Mac Os Catalina
Subsections
This tutorial demonstrates how to use the VMD plugin Timeline to analyzeand identify events in molecular dynamics (MD) trajectories. Timeline createsan interactive 2D box-plot - time vs. structural component - that can showdetailed structural events of an entire system over an entire MD trajectory.Events in the trajectory appear as patterns in the 2D plot.The plugin provides several built-in analysis methods, and the means todefine new analysis methods. Timeline can read and write data sets, allowingexternal analysis and plotting with other software packages. Timeline includesfeatures to help analysis of long trajectories and trajectories with largestructures.
In the main 2D box-plot graph, users identify events by looking for patterns ofchanging values of the analyzed parameter. The user can visually identifyregions of interest - rapidly changing structure values, clusters of brokenbonds, differences between stable and non-stable values, and similar. The usercan explore the resulting structures by tracing the mouse cursor(``scrubbing') over the identified areas. The structure is highlighted andthe trajectory is moved in time to track the highlight.
If you have any questions or comments on this tutorial, please email theTCB Tutorial mailing list at tutorial-l@ks.uiuc.edu. The mailing list is archived at http://www.ks.uiuc.edu/Training/Tutorials/mailing_list/tutorial-l/.
Getting Started

If you downloaded the tutorial from the web, the files that you will be needing can be found in a directory called timeline-tutorial-files. If you received the files through a workshop, they can be found in the same directory under the path /Workshop/timeline-tutorial.
- Unix/Mac OS X Users:In a Terminal window type:
cd <path to timeline-tutorial-files directory>
You can list the content of this directory, by using the command ls.
- Windows Users:Navigate to the timeline-tutorial-files directory using Windows Explorer.
Broken Timeline - Prologue Mac Os Download
This will place you in the directory containing all the necessary files. In the figure below, you can see the structure of this directory.
Required Programs
The following programs are required for this tutorial:- VMD: The tutorial assumes that you already have a working knowledge of VMD, which is available at http://www.ks.uiuc.edu/Research/vmd/(for all platforms)
- The VMD tutorial is available at
http://www.ks.uiuc.edu/Training/Tutorials/vmd/tutorial-html/
- The VMD tutorial is available at
- a text editor of your choice (we offer a few easy-to-use recommendations):
- UNIX: nedit (www.nedit.org)
- Windows XP: WordPad (included with OS). We recommend using WordPad as opposed to NotePad for this tutorial. However, please ensure that you save any files in this tutorial as .txt format files, as opposed to .rtf or .doc files, and to use the file extensions specified in the exercise, e.g. .tml.
- Mac OS X: Smultron (smultron.sourceforge.net), TextEdit (included with OS)
- a command prompt, such as a terminal in UNIX, Terminal.app in Mac OS X, or the DOS command prompt in Windows. Windows users can obtain a command prompt by clicking Start Programs Accessories Command Prompt.
Timeline in action: examine events in titin domain extension
Thissubsection provides a short run-through of of Timeline features to provide ageneral idea of how the plugin is used. Later sections will introduce thedisplay and user interface in greater detail. Here, we look at an MDtrajectory of the I91 domain of the muscle protein titin. In the simulationtrajectory, an external force is applied to the I91 domain with the SteeredMolecular Dynamics method: one terminus of the domain is fixed and a harmonicrestraint moving with constant velocity is applied to the other terminus,causing the domain to extend, unfold, and unravel. The secondary structurechanges greatly as the domain unfolds; examining these changes will give us someinitial insight into how the domain architecture responds to applied force.With the titin I91 extension trajectory loaded into VMD, the user starts Timeline byselecting Extension Analysis Timeline fromthe main VMD window, then generates the secondary structure plot by selectingCalculate Calc. Sec. Structure in the VMDTimeline window.
We examine trajectory events which appear as changes in value in the 2D plot.As the user moves the highlight cursor around the area of an event in the 2Dplot, she can explore the behavior of the 3D structure.Figure 2 illustrates examining an event in a Timelineplot of per-residue secondary structure. By moving the cursor around the eventarea indicated with a purple oval, the user observes a beta strand, depicted inyellow, losing its secondary structure as it peels away from another betastrand. Note that the purple oval is added for the purposes of this tutorial,the user must normally visually identify the event area. Here the event tonotice is the abrupt transition of a horizontal region of yellow to white: partof the domain losing the beta strand character (yellow) it had since the startof the trajectory and becoming random coil (white) as the domain extensionproceeds. A user would scrub the mouse highlight cursor around the area of thepurple oval, would produce the state shown inFigure 2a, before the local structure change, andexamine the steps needed to reach Figure 2b, after thelocal structure change has taken place. To ``scrub', click down the leftmouse button with the cursor in the area of interest in the 2D graph, then movethe cursor around the area of interest with the button still depressed - whilesplitting attention between the 2D graph, the changing 3D structure, and thechanging numerical values displayed in the Highlight Details panel (describedbelow).
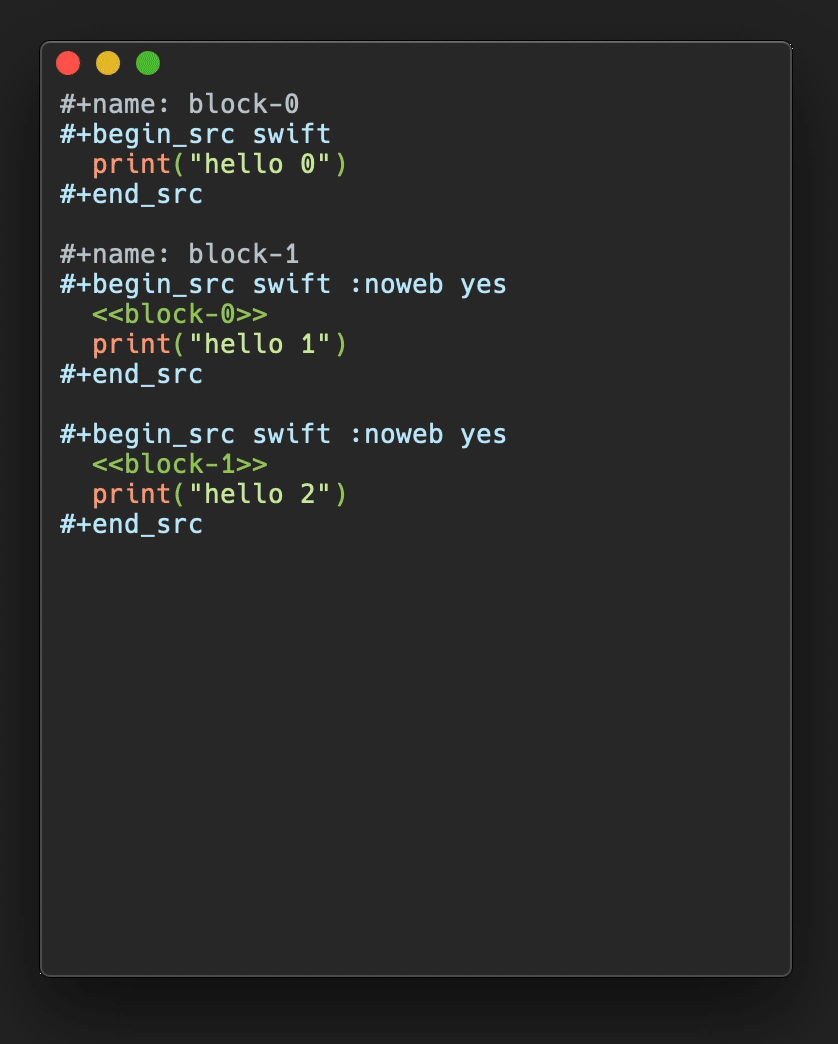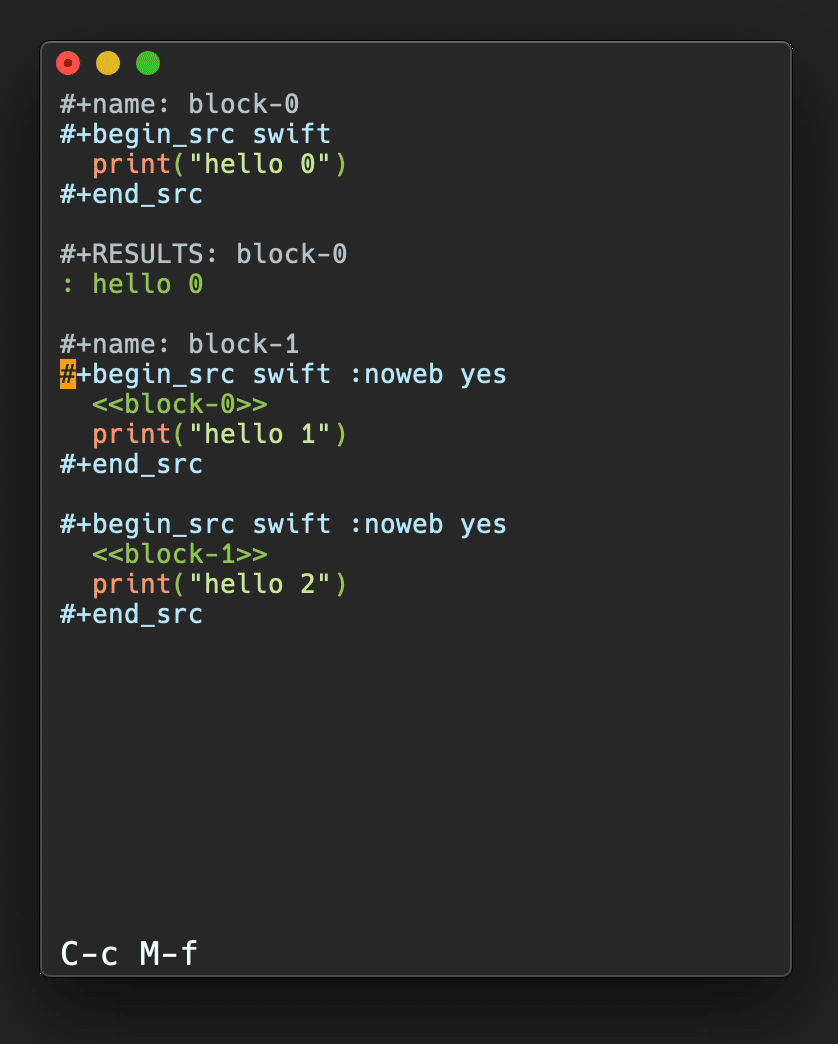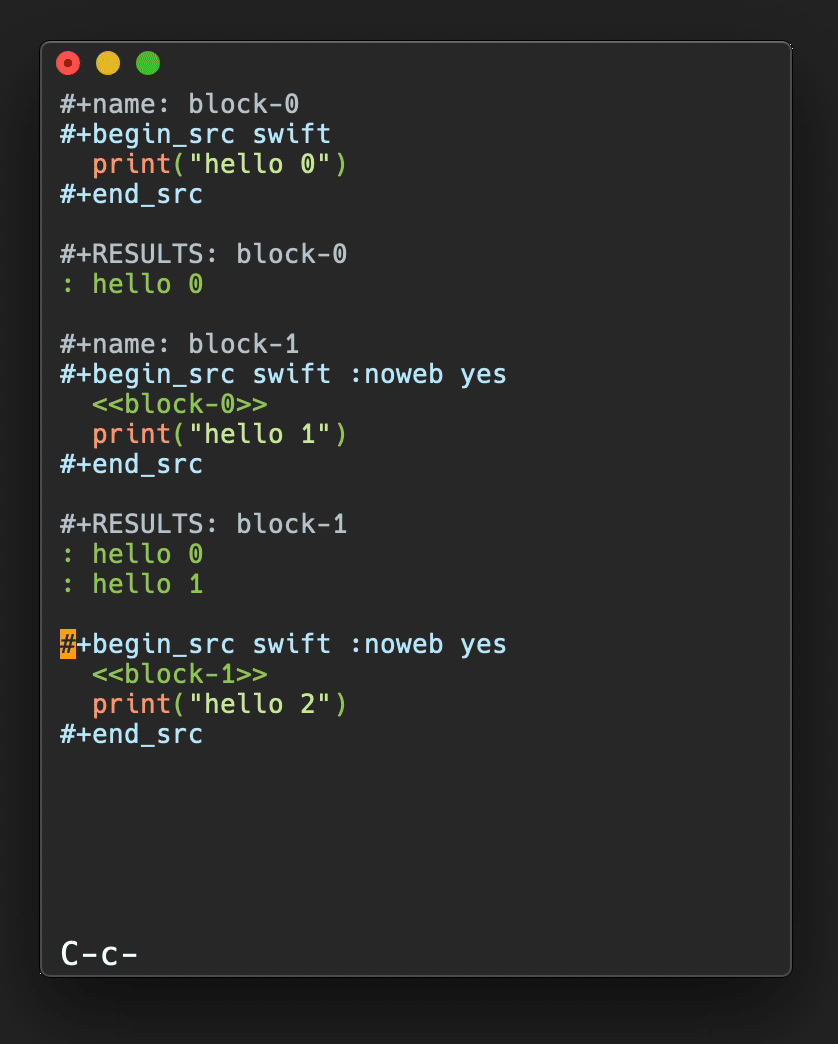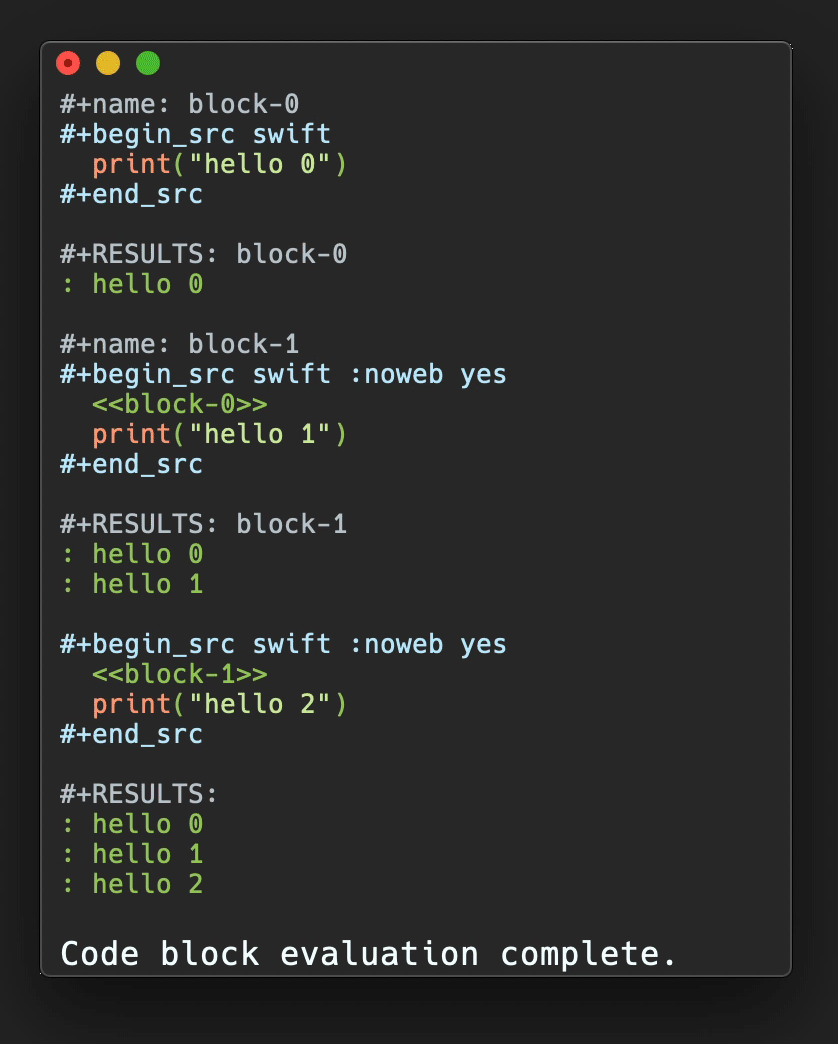
In Figure 3, a plot of salt bridge lifetimes during thetitin domain extension trajectory shown in Figure 2is displayed, illustrating moving the highlight cursor to examine a salt bridgebreaking event. This plot is per-selection; each selection is the pair ofresidues involved in a salt bridge. Only residue pairs that form a salt bridgefor at least one frame of the trajectory are listed. A highlighted salt bridgeis seen breaking between Figure 3a and3b. The Timeline highlighted representation can bemanipulated like any other VMD representation; here coloring of the highlightwas changed from the default ColorID 1/red to ResType toshow the different residue types of the salt bridge partners, and easily tracksalt bridge partners as they move apart.
In Figure 4 Timeline's Threshold Count plot is used to help find asuitable color scaling for the plot shown (per-residue Room Mean SquareFluctuation (RMSF)). The Threshold Count is the number of residues or selections in each frame with a value within a given range. As the Threshold Count range controls are adjusted as shown in Figure 4b, a patternwith multiple peaks appears (Figure 4c), this range is thenused to change the color scale, with the result shown in Figure4d. The Threshold Count sliders allow the users to veryquickly scan through threshold ranges, without taking the time to redraw theentire 2D plot, while still giving an overview of theentire trajectory.
Per-residue vs. per-selection calculations
Timeline plots data about a list of structural elements vs. time. There are two ways of defining the list of structural elements to be analyzed in Timeline.Per-residue: a list of residues; the protein and nucleotide residues in the user-defined selection. The default selection is ``all', so this defaults to being a list of all residues in the current molecule.
Per-selection: a list of any VMD selections. For example, a list ofsalt bridge pairs, a list of the segments in a large protein complex, a list of of favoritestructures in a molecule. These may be defined in built-in functions, listedexplicitly, or defined algorithmically in user scripts.
Next:Interface and controls Up:Timeline Tutorial Previous:Contents